Avian influenza surveillance in wild birds using different diagnostic techniques in Costa Rica
Abstract
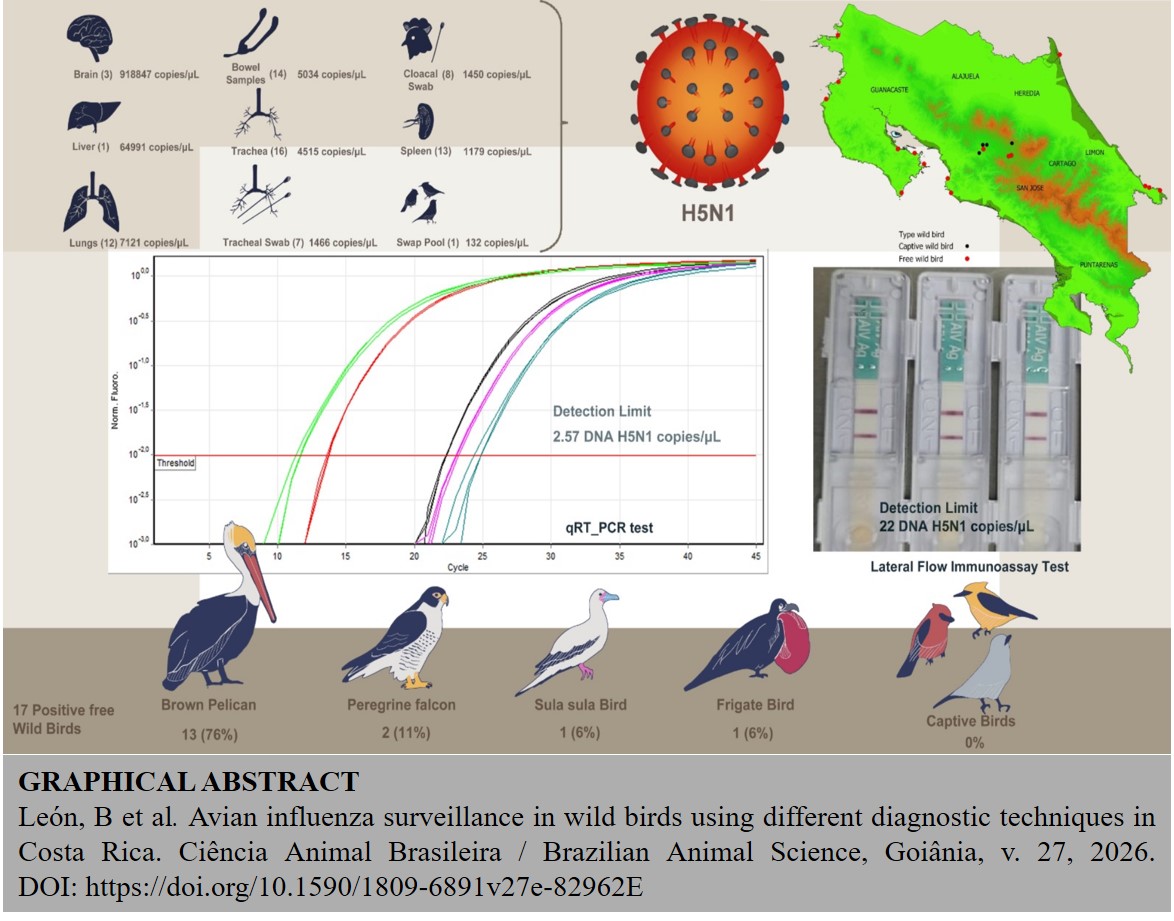
Avian influenza virus (AIV) surveillance is essential for monitoring outbreaks in wild bird populations. This study evaluated the performance of a rapid immunochromatographic test (RIT) compared to real-time PCR (qRT-PCR) and the agar gel immunodiffusion (AGID) test for AIV detection. RIT demonstrated greater sensitivity than AGID, detecting antigens at dilutions up to 1:512, whereas AGID only reached 1:16. At 1:512 dilution, qRT-PCR confirmed RIT results with a Ct value of 34.5, equivalent to 22 viral DNA copies/µL. The qRT-PCR detection limit for the AIV matrix gene was 2.57 DNA copies/µL. A total of 61 wild birds were reported to the National Animal Health Service in Costa Rica in 2023. Of these, 47 (77.0%) exhibited clinical signs of avian influenza, while 14 (23.0%) were dead. Thirteen taxonomic orders and 22 bird species were identified. with 32.2% of the samples belonging to Pelecaniformes. Using qRT-PCR, 17 birds (27.9%) tested positive for influenza A H5N1 virus. 76.5% were Pelecanus occidentalis, 11.8% Falco peregrinus, 5.9% red-footed boobies (Sula sula), and 5.9% great frigatebirds (Fregata minor). Fourteen (7 positive and 7 negative) were tested by RIT and qRT-PCR, showing 100% concordance between the two methods. Brain tissue showed the highest viral load (918,847 copies/µL), while cloacal swabs had the lowest (1,450 copies/µL). Tracheal samples were most frequently submitted (94.1%) and had a 93.8% positive rate. Geospatial analysis revealed most positive cases were from the Pacific Coast. Organ-specific testing highlighted the risk of false negatives, emphasizing the need for optimized tissue sampling. Additionally, 200 cloacal swabs from birds in four rescue centers in Costa Rica’s Central Valley tested negative by RIT. These findings support the complementary use of RIT with qRT-PCR for effective field and laboratory AIV surveillance. Monitoring and improved diagnostic strategies are critical for early detection and control of outbreaks. in Costa Rica.
Keywords: Zoonosis; avian; H5N1; wild birds.
Downloads
References
1. Hill NJ, Bishop MA, Trovão NS, Ineson KM, Schaefer AL, Puryear WB, et al. Ecological divergence of wild birds drives avian influenza spillover and global spread. PLoS Pathog. 2022;18:e1010062. Available at: https://doi.org/10.1371/journal.ppat.1010062
2. Venkatesh D, Poen MJ, Bestebroer TM, Scheuer RD, Vuong O, Chkhaidze M, et al. Avian influenza viruses in wild birds: virus evolution in a multihost ecosystem. J. Virol. Amer. Soc. Microbiol. 2018;92:10.1128/jvi.00433-18. Available at: https://doi.org/10.1128/jvi.00433-18
3. Gonzalez-Reiche AS, Müller ML, Ortiz L, Cordón-Rosales C, Perez DR. Prevalence and diversity of low pathogenicity avian influenza viruses in wild birds in Guatemala, 2010-2013. Avian Dis. 2016;60:359–364. Availble at: https://doi.org/10.1637/11130-050715-Reg
4. Fujimoto Y, Usui T, Ito H, Ono E, Ito T. Susceptibility of wild passerines to subtype H5N1 highly pathogenic avian influenza viruses. Avian Pathol. J, WVPA. 2015;44:243–247. Available at: https://doi.org/10.1080/03079457.2015.1043235
5. Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, et al. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. Available at: https://doi.org/10.1371/journal.ppat.0030061
6. Knight-Jones TJD, Hauser R, Matthes D, Stärk KDC. Evaluation of effectiveness and efficiency of wild bird surveillance for avian influenza. Vet. Res. 2010;41:50. Available at: https://doi.org/10.1051/vetres/2010023
7. Perera CL, Díaz de Arce H, Pérez LJ. Actualización y perspectivas en el diagnóstico del virus de la influenza aviar. Rev. Salud Anim. 2011;33:01–7. Available at: https://www.produccion-animal.com.ar/produccion_aves/enfermedades_aves/112-influenza_actualizacion.pdf
8. Wibowo MH, Untari T, Artanto S, Putri K, Amanu S, Asmara W. Evaluation of rapid detection kit against avian influenza A virus and H5 subtype for field sample. Indones. J. Biotechnol. 2016;21:48–55. Available at: https://doi.org/10.22146/ijbiotech.26792
9. Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, et al. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. Available at: https://doi.org/10.1128/JCM.40.9.3256-3260.2002
10. León B, Chaves G, Koster LG, Jenkins-Moore M, Carrillo C, Méndez D. Isolation and identification of pandemic influenza virus H1N1 / 2009 S-OIV from commercial and backyard swine in Costa Rica. Rev. Cienc. Vet. 2011;29:53–81.
11. Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005;109:365–379. Available at: https://doi.org/10.1042/CS20050086
12. Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinform. 2006;7:85. https://doi.org/10.1186/1471-2105-7-85
13. Nassiri M, Torshizi, ME, Ghovvati S, Doosti M. Evaluation of different statistical methods using SAS software: an in silico approach for analysis of real-time PCR data. J. Appl. Stat. 2018;45:306–319. Available at: https://doi.org/10.1080/02664763.2016.1276890
14. Gall A, Hoffmann B, Harder T, Grund C, Beer M. Universal primer set for amplification and sequencing of HA0 cleavage sites of all influenza A viruses. J. Clin. Microbiol. 2008;46:2561–2567. Available at: https://doi.org/10.1128/JCM.00466-08
15. Zhou B, Donnelly ME, Scholes DT, George KS, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J. Virol. 2009;83:10309–10313. Available at: https://doi.org/10.1128/jvi.01109-09
16. Chua T-H, Ellis TM, Wong CW, Guan Y, Ge SX, Peng G, et al. Performance evaluation of five detection tests for avian influenza antigen with various avian samples. Avian Dis. 2007;51:96–105. Available at: https:// DOI.org/ 10.1637/0005-2086(2007)051[0096:PEOFDT]2.0.CO;2
17. Slomka MJ, Pavlidis T, Banks J, Shell W, McNally A, Essen S, et al. Validated H5 Eurasian real-time reverse transcriptase-polymerase chain reaction and its application in H5N1 outbreaks in 2005-2006. Avian Dis. 2007;51:373–377. Available at: https://doi.org/10.1637/7664-060906R1.1
18. Spackman E, Pedersen JC, McKinley ET, Gelb J. Optimal specimen collection and transport methods for the detection of avian influenza virus and Newcastle disease virus. BMC Vet. Res. 2013;9:35. Available at: https://doi.org/10.1186/1746-6148-9-35
19. McDuie F, T. Overton C, A. Lorenz A, L. Matchett E, L. Mott A, A. Mackell D, et al. Mitigating risk: predicting H5N1 avian influenza spread with an empirical model of bird movement. Transbound Emerg Dis. 2024;2024:5525298. Available at: https://doi.org/10.1155/2024/5525298
20. Lebarbenchon C, Sreevatsan S, Ramakrishnan MA, Poulson R, Goekjian V, Di Matteo JJ, et al. Influenza A viruses in American white pelican (Pelecanus erythrorhynchos). J. Wildl. Dis. 2010;46:1284–1289. Available at: https://doi.org/10.7589/0090-3558-46.4.1284
21. Adzic B, Goletic S, Pejoviс N, Vizi A, Yolshin N. First case of highly pathogenic avian influenza H5N1 in Montenegro. Acta Vet.-Beogr. 2024;74:145–158. Available at: https://doi.org/10.2478/acve-2024-0011
22. Alexandrou O, Malakou M, Catsadorakis G. The impact of avian influenza 2022 on Dalmatian pelicans was the worst ever wildlife disaster in Greece. Oryx. 2022;56:813–813. Available at: https://doi.org/10.1017/S0030605322001041
23. Stoimenov GM, Goujgoulova GV, Nikolov B, Petrova R, Teneva A, Dimitrova I. Histopathological findings in Dalmatian pelicans (Pelecanus crispus) naturally infected with avian influenza subtype A H5N1 in Bulgaria. J. Hell. Vet. Med. Soc. 2018;68:369–376. Available at: https://doi.org/10.12681/jhvms.15493
24. Cardona CJ, Xing Z, Sandrock CE, Davis CE. Avian influenza in birds and mammals. Comp. Immunol. Microbiol. Infect. Dis. 2009;32:255–273. Available at: https://doi.org/10.1016/j.cimid.2008.01.001
25. Lara Rivera DA. Determinación de anticuerpos circulantes contra el virus de influenza aviar en aves del Centro de Rehabilitación de Vida Silvestre ARCAS, Flores, Petén, Guatemala. [Thesis]: Guatemala: Universidad de San Carlos; 2008. Available at: http://www.repositorio.usac.edu.gt/7406/
26. Neu Toscano AE. Determinación de anticuerpos circulantes contra el virus de influenza aviar en aves del Parque Zoológico Minerva de Quetzaltenango, Guatemala. [Thesis]: Guatemala: Universidad de San Carlos; 2016. Available at: http://biblioteca.usac.edu.gt/tesis/10/10_1589.pdf
27. Slusher MJ, Wilcox BR, Lutrell MP, Poulson RL, Brown JD, Yabsley MJ, et al. Are passerinae bords reservoirs for influenza A viruses? J. Wildl. Dis. 2014;50:792–809. Available at: https://doi.org/10.7589/2014-02-043
28. Brown JD, Berghaus RD, Costa TP, Poulson R, Carter DL, Lebarbenchon C, et al. Intestinal excretion of a wild bird-origin H3N8 low pathogenic avian influenza virus in mallards (Anas platyrhynchos). J. Wildl. Dis. 2012;48:991–998. Available at: https://doi.org/10.7589/2011-09-280
29. Swayne DE, Slemons RD. Using mean infectious dose of high- and low-pathogenicity avian influenza viruses originating from wild duck and poultry as one measure of infectivity and adaptation to poultry. Avian Dis. 2008;52:455–460. Available at: https://doi.org/10.1637/8229-012508-Reg.1
Downloads
Additional Files
Published
How to Cite
Issue
Section
License
Copyright (c) 2026 Brazilian Animal Science/ Ciência Animal Brasileira

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
- Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
- Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
- Authors are permitted and encouraged to post their work online (e.g. in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See The Effect of Open Access).
Data statement
-
The research data is contained in the manuscript



























