DOI: 10.1590/1089-6891v20e-47449
MEDICINA VETERINÁRIA
DIARRHEAGENIC Escherichia coli IN RAW MILK, WATER, AND CATTLE FECES IN NON-TECHNIFIED DAIRY FARMS
Escherichia coli DIARREIOGÊNICA EM LEITE CRU, ÁGUA E FEZES BOVINAS DE PROPRIEDADES LEITEIRAS NÃO TECNIFICADAS
Laryssa F. Ribeiro1* ORCID – http://orcid.org/0000-0003-1943-7897
Mayhara M. C. Barbosa² ORCID – http://orcid.org/0000-0002-1970-8266
Fernanda R. Pinto³ ORCID – http://orcid.org/0000-0002-0794-1984
Leticia F. Lavezzo⁴ ORCID – http://orcid.org/0000-0002-1461-6362
Gabriel A. M. Rossi⁵ ORCID – http://orcid.org/0000-0001-7967-7628
Henrique M. S. Almeida⁴ ORCID – http://orcid.org/0000-0002-7631-4271
Luiz A. Amaral⁴ ORCID – http://orcid.org/0000-0002-5297-4961
¹Centro Universitário Mário Palmério (UNIFUCAMP), Monte Carmelo, MG, Brazil
²Instituto Federal de Educação, Ciência e Tecnologia do Ceará (IFCE), Quixadá, CE, Brazil.
³Universidade Federal de Pelotas (UFPEL), Pelotas, RS, Brazil.
⁴Universidade Estadual Paulista, Faculdade de Ciências Agrárias e Veterinárias, Jaboticabal, SP, Brazil.
⁵ Centro Universitário Central Paulista (UNICEP), São Carlos, SP, Brazil.
*Correspondent author - laryssaribeiro84@gmail.com
Abstract
This study focused on detecting diarrheagenic Escherichia coli, enteropathogenic E. coli (EPEC), Shiga-toxin-producing E. coli (STEC), enterohemorrhagic E. coli (EHEC or STEC:EPEC), enterotoxigenic E. coli (ETEC), and enteroaggregative E. coli (EAEC) in raw milk, water, and cattle feces sampled from non-technified dairy farms located in the northeastern São Paulo State, Brazil. Thirty-six water samples were collected at different points, namely, water wells (8 samples), water intended for human consumption (8 samples), water from milking parlor (8 samples), and water intended for animal consumption (7 samples), headwaters (1 sample), rivers (3 samples), and reservoirs (1 sample). Three raw milk samples were taken directly from bulk tanks in each farm, totalizing 24 samples. Feces samples were collected using rectal swabs from 160 bovines (20 animals per farm). E. coli was detected in 128 feces samples (80%), 16 raw milk samples (66.67%), and 20 water samples (55.56%). STEC (26 samples, 16.25%), EPEC (10 samples, 6.25%), STEC: EPEC (5 samples, 3.13%), and STEC: ETEC (1 sample, 0.63%) were the most prevalent strains detected in samples from cattle feces. EPEC, STEC, and STEC: EPEC strains were detected in 4.17% (1 sample), 16.67% (4 samples), and 4.17% (1 sample) of raw milk samples, respectively. STEC strains were detected in water used in the milking parlor, while no EAEC strain was detected. As a conclusion, cattle feces are important contamination sources of pathogenic E. coli in non-technified dairy farms and, consequently, cross-contamination among feces, water, and/or raw milk can occur. The use of quality water and hygienic practices during milking are recommended to avoid contamination since pathogens can be transmitted to humans via raw milk or raw milk cheese ingestion.
Keywords: EAEC, ETEC, EPEC, STEC, public health
Resumo
Este estudo teve como objetivo realizar a detecção de Escherichia coli diarreiogênica (EPEC, STEC, ETEC e EAEC) em leite, água e fezes bovinas em pequenas propriedades leiteiras localizadas na Região Nordeste do Estado de São Paulo, Brasil. E. coli foi detectada em amostras obtidas de fezes (80%), leite cru (66,67%) e água (55,56%). STEC, EPEC, STEC:EPEC e STEC:ETEC foram as cepas mais prevalentes em amostras de fezes bovinas, respectivamente. Em relação ao leite cru, cepas de EPEC, STEC e STEC:EPEC foram detectadas em 4,17%, 16,67% e 4,17% das amostras, respectivamente. Ainda, detectou-se STEC na amostra de água utilizada na sala de ordenha, enquanto EAEC não foi detectada em nenhuma amostra. Conclui-se que fezes de bovinos é uma importante fonte de contaminação de E. coli patogênicas em propriedades leiteiras e podem consequentemente contaminar o leite cru e água. A importância da qualidade da água e da adoção efetiva de práticas higiênicas durante a obtenção do leite para evitar a contaminação são recomendadas devido à possibilidade de transmissão de microorganismos patogênicos a seres humanos devido a ingestão de leite cru ou queijos produzidos a partir de leite não pasteurizado.
Palavras-chave: EAEC, ETEC, EPEC, STEC, saúde pública
Received on: June, 17th, 2017.
Accepted on: June, 10th. 2019.
Introduction
Waterborne disease outbreaks are major challenges for public health worldwide(1). Developing countries are more susceptible to such epidemics due to their poorer sanitation control and water quality than developed countries, which are most affected by emerging water-treatment resistant pathogens, technical failures, or even inadequate control measures(2).
The presence of pathogenic Escherichia coli strains is an important indicator of public health hazards. These bacteria are divided into six diarrheagenic groups: enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), and diffusely adherent E. coli (DAEC)(3). EPEC is a common cause of childhood diarrhea in developing countries due to its ability to adhere to the intestinal mucosa, resulting in nutrient malabsorption(4). The formation of A/E lesions causes a decrease in the absorption capacity of the intestinal mucosa, which leads to a rupture of the electrolytic balance and hence to diarrhea(5). Diarrhea caused by EPEC probably results from multiple mechanisms, including ion secretion, increased intestinal permeability, intestinal inflammation, and loss of absorption surface(6). STEC can produce toxins that cause illnesses if ingested, such as hemorrhagic colitis and hemolytic uremic syndrome (HUS)(4). These toxins, also known as verotoxins, produce a distinct and irreversible cytotoxic effect on Vero cells(7).
ETEC toxins induce fluid secretion, causing childhood diarrhea. Their outbreaks are frequently associated with poor handling or poor water quality and hence contaminated food consumption(4). These microorganisms colonize intestinal mucosa surface and release LT (thermolabile) and ST (thermostable) enterotoxins, causing diarrhea(8). In addition, they express surface adhesins which allow the colonization of intestinal epithelial cells(9). EAEC is also a cause of persistent diarrhea in children and can lead to chronic inflammation. EAEC pathogenesis is determined by the organism's ability to adhere to intestinal cells, produce enterotoxins and cytotoxins, and induce inflammation. Dispersin, which is antigenic antiaggregatory protein encoded by the aap gene in the pAA plasmid and regulated by aggR, modulates fimbrial adhesion and facilitates penetration of the microorganism through intestinal mucus by binding to lipopolysaccharide and altering the electrostatic properties of the EAEC outer membrane surface(10).
Some pathogenic E. coli strains are adapted to survive in aquatic environments and associated with high-mortality in waterborne outbreaks. Diarrheagenic strains were detected in some water sources such as raw water, surface water, animal/human wastewater, irrigation water, swimming water, and municipal water contaminated with feces. Thus, preventing the contamination of public water supplies and proper disinfection methods such as chlorine and ultraviolet light treatments are recommended(11).
Raw milk and dairy products are usually associated with diarrheagenic E. coli contamination(12). Some diarrheagenic E. coli outbreaks due to the consumption of unpasteurized milk are described in the literature(13,14). Furthermore, high prevalence of potentially pathogenic E. coli strains in raw milk and raw milk cheeses is reported in several countries worldwide(15).
Contamination sources of diarrheagenic E. coli in dairy products are poorly treated water or even direct contact with cattle feces. Vicente et al.(9) detected a prevalence of 100% on dairy herds and detected at least one animal containing STEC strains in feces. These authors highlighted bovines as important sources of environmental contamination, mainly during the rainy season. Furthermore, bovines are considered as the most important reservoirs of STEC and EPEC for humans, and transmission occurs via ingestion of contaminated food or water(16).
The detection of virulence factors such as stx1 (stx1A), stx2 (stx2A), intimin (eae), transcriptional activator AggR (aggR), heat-labile toxin type 1(eltB), heat-stable enterotoxin (estA, estB), and plasmid pCVD432 (aatA) in E. coli isolates has been recommended to evaluate potential risk for public health(17 - 22). Overall, this study focused on detecting the presence of diarrheagenic E. coli (EPEC, STEC, ETEC, and EAEC) in samples of milk, water, and cattle feces from dairy farms in the northeastern São Paulo State, Brazil.
Materials and methods
Sampling was performed in eight non-technified dairy farms located in Jaboticabal municipality, São Paulo State (Brazil), between 2010 and 2011. And by non-technified dairy farms, we mean properties with a maximum cattle population of 20 cross-breed animals, maximum milk production of 300 liters, no attending veterinarian, no sanitary control, and family production with family employees. Seven farms used hand milking and one mechanical milking. These farms have a daily production of about 250-260 liters of raw milk. Milking is performed twice a day (in the morning and afternoon).
Thirty-six water samples were collected at different sampling points in the farms, according to their own rearing conditions, such as from wells (8 samples), milking parlor (8 samples), headwater (1 sample), river (3 samples), and reservoir (1 sample), and water intended for human (8 samples) and animal (7 samples) consumptions. Three raw milk samples were collected directly from bulk tanks in each farm (at the beginning, middle, and end of milking), totalizing 24 samples. Water and milk samples were taken using sterilized glasses with a maximum capacity of 500 mL(23). Feces samples were collected using rectal swabs from 160 bovines (20 animals per farm)(24).
E. coli was isolated according to Vicente et al.(9). Three to five colonies were picked from each plate for biochemical identification(25). DNA from all isolates was extracted(26) and examined for quality and integrity by running on an agarose gel.
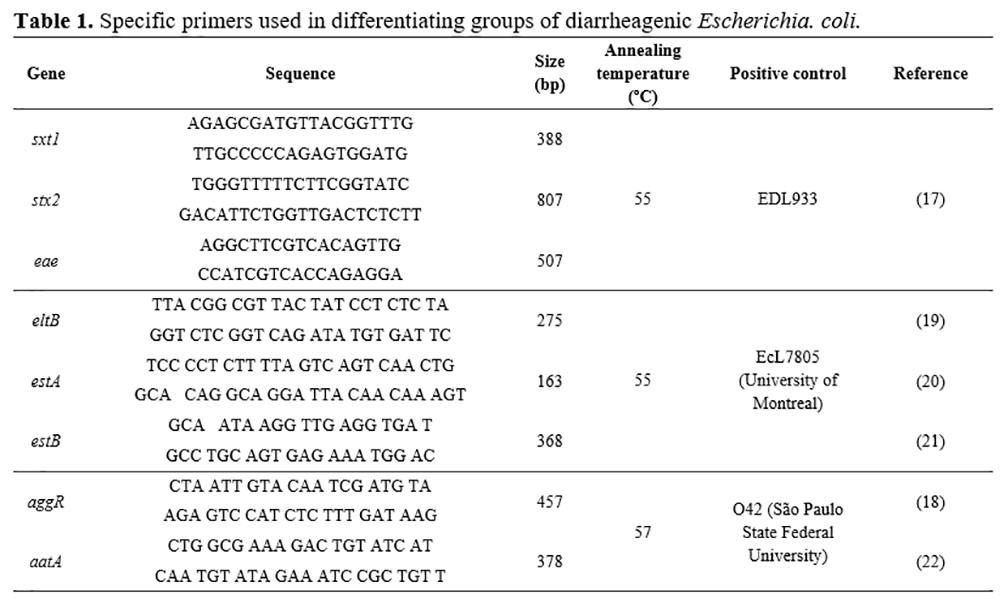
To detect diarrheagenic E. coli strains, E. coli isolates were screened for virulence genes, using three conventional multiplex PCR with a set of specific primers for amplification. A multiplex PCR was performed to detect EPEC and STEC. For EPEC, it was detected eae gene, and for STEC genes stx1 and stx2. Another multiplex PCR was done for ETEC genes, and the genes eltB, estA, and estB were detected. Moreover, a third multiplex PCR was performed for EAEC, and the genes aggR and aatA were detected. Table 1 describes the genes, sequences, sizes (bp), annealing temperatures (ºC), and strain positive controls.
Results and discussion
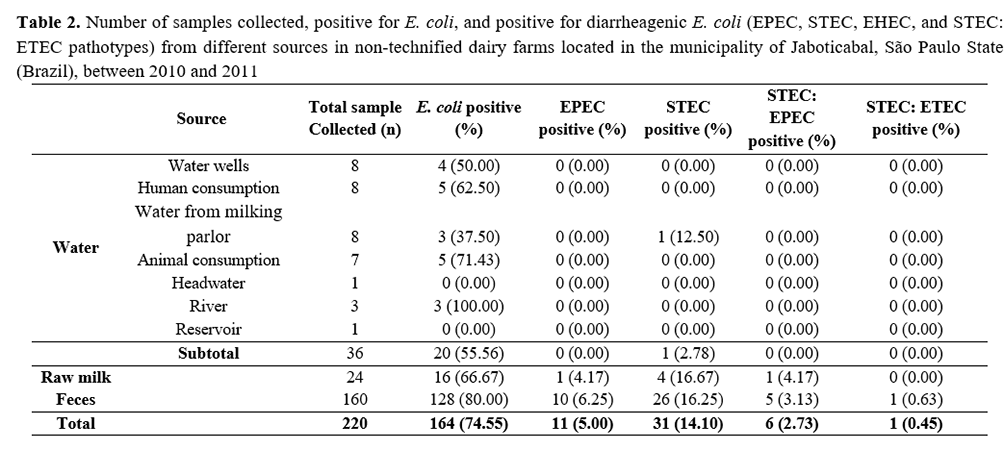
Out of the 220 samples collected, E. coli was isolated from 164 samples (74.55%), as shown in Table 2. This bacterium was detected with higher prevalence in samples of feces (80.00%), raw milk (66.67%), and water sources (55.56%). It was detected in samples of water for human consumption (62.50%), demonstrating a possible risk to public health due to the transmission of waterborne diseases. Furthermore, the presence of STEC in water from water milking parlor demonstrates a risk of raw milk contamination with major pathogens during milking if hygienic practices are not properly carried out(27).
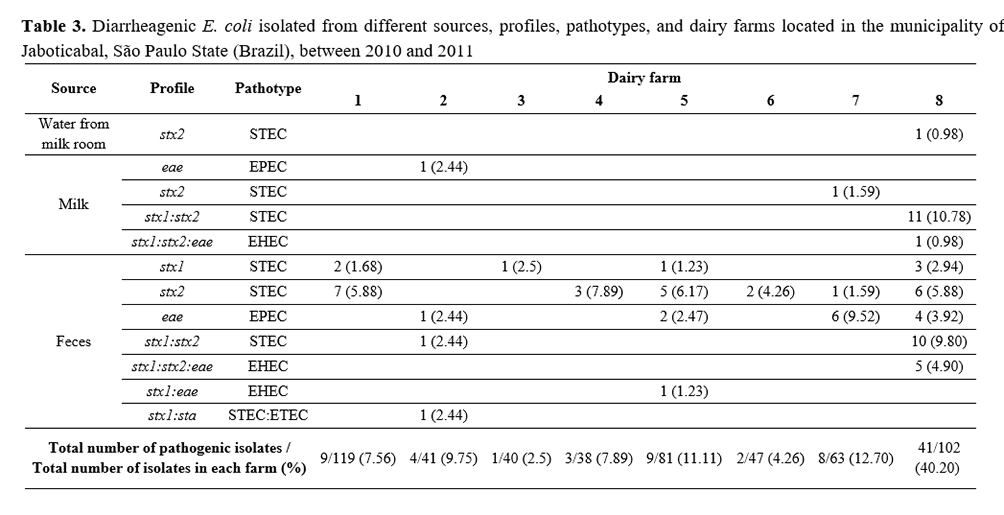
Five hundred and thirty-one E. coli isolates were obtained: 67 from water, 392 from feces, and 72 from milk samples. All farms (100%) had at least one potentially pathogenic isolate (Table 3). An atypical pathotype containing stx1: sta genes was detected in one bovine feces sample.
Ingestion of raw milk or raw milk cheeses can pose risks to public health(15) since we detected EPEC (4.17%), STEC (16.67%), and EHEC (4.17%) strains in raw milk. Furthermore, pathogenic strains were detected in cattle feces, highlighting bovine as an important reservoir of such strains, thus being a potential source of contamination of milk and other dairy products(9, 28). There was a great prevalence of STEC, EPEC, EHEC, and STEC: ETEC in cattle feces.
E. coli strains belonging to EAEC group were isolated from food and stool samples of diarrheagenic patients in Colombia, demonstrating the importance of this foodborne pathogen in developing countries(29). No EAEC strains were detected in the samples evaluated in the present study, which could be explained by the fact that animals are not commonly reservoirs for EAEC strains(30).
Similar high frequencies of diarrheagenic strains have also been reported in other Brazilian dairy farms. Sandrini et al.(31) detected the presence of STEC strains in 95% of dairy farms evaluated in the municipality of Pelotas, Rio Grande do Sul State (Brazil). Moreover, Irino et al.(32) observed STEC isolation rates ranging from 3.8 to 84.6% in six dairy farms of São Paulo State (Brazil).
Farrokh et al.(28) stated that milk and other dairy products are important contamination sources of STEC strains causative of outbreaks, as demonstrated in our study. Pathogen surveillance is a risk management measure for public health protection(33), in which it is highlighted the need for long-term strategies to ensure safe dairy products(34). Also, there has been reported a regular consumption of raw milk and other dairy products in Brazil(35).
In this study, the seven farms (1, 2, 3, 4 5, 6, and 7) showed low percentages of pathogenic E. coli strains, but only farm 8 had 40.20% of the total isolates (41/102) (Table 3). This farm had a high occurrence of isolates containing the gene stx2 in milk and feces samples, and one isolate in water samples. In this property, there might have been cross-contamination among animal feces, milking parlor water, and raw milk due to high environmental contamination, demonstrating the need for hygienic practices(36).
Cattle feces are a common source of pathogenic E. coli (STEC and EPEC) as demonstrated in the present study. Vicente et al.(9) evaluated the frequency of stx1, stx2 and eae genes in dairy farms and observed high prevalence (72.16%) of the gene stx in isolates from feces samples of asymptomatic animals. Thus, teats and udders can be contaminated with infected feces and hence contaminate raw milk in case of unsatisfactory hygienic conditions during milking(37) or by mastitic milk(38).
Raw milk from bulk tanks can pose a risk of transmission of STEC strains to consumers(39), just as other dairy products, which could threaten public health if contaminated with E. coli. Outbreaks can be caused by pathogenic E. coli when consuming cheese made of raw milk(13, 14, 40, 41).
Paneto et al.(42) evaluated 50 samples of raw cheeses from supermarkets in the Brazilian midwestern region and observed that 96% of them were contaminated with E. coli, posing a risk to the local population. Other authors also observed a higher risk of E. coli infection in areas where consumption of unpasteurized dairy products is common, such as the state of São Paulo(43), highlighting the importance of pasteurizing milk to assure public health(44).
Conclusion
Cattle feces were the most important sources of pathogenic Escherichia coli in the studied dairy farms and hence could contaminate raw milk and water. Therefore, water quality and effective hygienic practices during milking are crucial to avoid raw milk contamination. These conditions should be considered owing to the possibility of transmission of pathogenic microorganisms to humans via ingestion of raw milk or its derived cheeses.
References
1 Breathnach AS, Cubbon MD, Karunaharan RN, Pope CF, Planche TD. Multidrug-resistant Pseudomonas aeruginosa out- breaks in two hospitals: association with contaminated hospital waste-water systems. Journal of Hospital Infection. 2012; 82:19–24.
2 Man H, van den Berg, HHJL, Leenen EJTM, Schijven JF, Schets FM, van der Vliet, JC, van Knapen F, de Roda Husman AM. Quantitative assessment of infection risk from exposure to waterborne pathogens in urban floodwater. Water Research. 2014; 48:90–99.
3 Kaper JB, Nataro JP, Mobbley HLT. Pathogenic Escherichia coli. Nature Reviews. 2004; 2:123-140.
4 FDA, Food and Drug Administration. The Bad Bug Book. 2nd ed, 2012 292p. Available at: http://www.fda.gov/downloads/Food/FoodborneIllnessContaminants/UCM297627.pdf, Accessed on 14 dec. 2015.
5 Clarke SC, Haigh RD, Freestone PPE, Williams PH. Virulence of Enteropathogenic Escherichia coli, a Global Pathogen. Clinical Microbiology Reviews. 2003; 16(3):365–378.
6 Shaw RK, Cleary J, Murphy MS, Frankel G, Knutton S. Interaction of enteropathogenic Escherichia coli with human intestinal mucosa: role of effector proteins in brush border remodeling and formation of attaching and effacing lesions. Infection and Immunity. 2005; 73(2):1243–51.
7 Konowalchuk J, Speirs JI, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infection and Immunity. 1977; 18(3):775-779.
8 Nataro JP, Kaper J B. Diarrheagenic Escherichia coli. Clinical Microbiology Reviews. 1998; 11:142–201.
9 Vicente HIG, Amaral LA, Cerqueira AMF. Shigatoxigenic Escherichia coli Serogroups O157, O111 and O113 in feces, water and milk samples from dairy farms. Brazilian Journal of Microbiology. 2005; 36:217-222.
10 Okhuysen PC, Dupont HL. Enteroaggregative Escherichia coli (EAEC): A Cause of Acute and Persistent Diarrhea of Worldwide Importance. Journal of Infectious Diseases. 2010; 202:503-505.
11 Saxena T, Kaushik P, Mohan MK. Prevalence of E. coli O157:H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagnostic Microbiology and Infectious Disease. 2015; 82:249-264.
12 Serrano NS, Zweifel C, Corti S, Stephan R. Microbiological quality and presence of foodborne pathogens in raw milk cheeses and raw meat products marketed at farm level in Switerzland. Italian Journal of Food Safety. 2018: 7(2):7337.
13 Mylius M, Dressman J, Pulz M, Pallasch G, Beyrer K, Clauben K, Allerberger F, Fruth A, Lang C, Prager R, Flieger A, Schlager S, Kalhöfer D, Mertens E. Shiga toxin-producing Escherichia coli O103:H2 outbreak in Germany after school trip to Austria due to raw milk cow, 2017 – The important role of international collaboration for outbreak investigations. International Journal of Medical Microbiology. 2018; 308(5):539-544.
14 Jones G, Lefèvre S, Donguy MP, Nisavanh A, Terpant G, Fougere E, Vaissieri E, Guinard A, Mailles A, Valk H, Fila M, Tanné C, Borgne CL, Weill FX, Bonacorsi S, Silva NJ, Mariani-Kurkdjian. Outbreak of Shiga-producing Escherichia coli (STEC) O26 paediatric haemolytic uraemic syndrome (HUS) cases associated with the consumption of soft raw cow's milk cheeses, France, March to May 2019. Euro Surveillance. 2019; 24(22): pii=1900305
15 Omarak RA, Hinenoya A, Awastshi SP, Iguchi A, Shima A, Elbagory ARM, Yamasaki S. Prevalence and pathogenic potential of Escherichia coli isolates from raw milk and raw milk cheese in Egypt. International Journal of Food Microbiology. 2016; 221:69-76.
16 Salaheen S, Kim SW, Karns JS, Hovingh E, Haley BJ, Kessel JASV. Metagenomic analysis of the fecal microbiomes from Escherichia coli O157:H7 – shedding and non-shedding cows on a single dairy farm. Food Control. 2019; 102:76-80.
17 China B, Pirson V, Mainil J. Typing of Bovine Attaching and Effacing Escherichia coli by Multiplex In Vitro Amplification of Virulence-Associated Genes. Applied and Environmental Microbiology. 1996; 62:3462-3465.
18 Cerna JF, Nataro JP, Estrada-Garcia T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. Journal of. Clinical Microbiology. 2003; 41:2138-2140.
19 Furrer B, Candrian U, Luthy J. Detection and identification of E. coli producing heat-labile enterotoxin type I by enzymatic amplification of a specific DNA fragment. Letters in Applied Microbiology. 1990; 10:31-34.
20 Ngeleka M, Pritchard J, Appleyard G, Middleton DM, Fairbrother JM. Isolation and association of Escherichia coli AIDA-I/STb, rather than EAST1 pathotype, with diarrhea in piglets and antibiotic sensitivity of isolates. Journal of Veterinary. Diagnostic Investigation. 2003; 15:242–252.
21 Lortie LA, Dubreuil JD, Harel J. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. Journal of Clinical Microbiology. 1991; 29(3):656–659.
22 Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H.. Development of PCR for screening of enteroaggregative Escherichia coli. Journal of Clinical Microbiology. 1996; 33:701-705.
23 APHA. American Public Health Association. Standard methods for the examination of water and wastewater, AWWA, 20th, 1998.
24 Cerqueira AMF, Guth BEC, Joaquim RM, Andrade JRC. High occurrence of Shiga toxin-producing Escherichia coli (STEC) in healthy cattle in Rio de Janeiro State, Brazil. Veterinary Microbiology. 1999; 70, 111-121.
25 Koneman EWA, SD, Janda WM, Schreckenberger PC, Winn JWC. Diagnóstico Microbiológico. Medsi, 5ed., 2001; 177-261
26 Keskimaki M, Eklund M, Pesonen H, Heiskanen T, Siitonen A. EPEC, EAEC and STEC in stool specimens: Prevalence and molecular epidemiology of isolates. Diagnostic Microbiology and Infectious Disease. 2001; 40:151-156
27 Martin A, Beutin L. Characteristics of Shiga toxin-producing Escherichia coli from meat and milk products of different origins and association with food producing animals as main contamination sources. International Journal of Food Microbiology. 2011; 146:99-104.
28 Farrokh C, Jordan K, Auvray F, Glass K, Oppegaard H, Raynaud S, Thevenot D, Condron R, De Reu K, Govaris A, Heggum K, Heyndrickx M, Hummerjohann J, Lindsay D, Miszczycha S. (2013). Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. International Journal of Food Microbiology. 2013; 162:190–212
29 Rúgeles LC, Bai J, Martínez AJ, Vanegas MC, Gómez-Duarte OG. Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. International Journal of Food Microbiology. 2010; 138:282 -286
30 Uber AP, Trabulsi LR, Irino K, Beutin L, Ghilardi ACR, Gomes TAT, Liberatore AMA, Castro AFP, Elias WP. (2006). Enteroaggregative Escherichia coli from humans and animals differ in major phenotypical traits and virulence genes. FEMS Microbiology Letters. 2006; 256:251-257.
31 Sandrini CNM, Pereira MA, Brod CS, Carvalhal JB, Aleixo JAG. Escherichia coli verotoxigênica: isolamento e prevalência em 60 propriedades de bovinos de leite da região de Pelotas, RS, Brasil. Ciência Rural. 2007; 37:175-182.
32 Irino K, Kato MAMF, Vaz TMI, Ramos II, Souza MAC, Cruz AS, Gomes TAT, Vieira MAM, Guth BEC. Serotypes and virulence markers of Shiga toxin-producing Escherichia coli (STEC) isolated from dairy cattle in São Paulo State, Brazil. Veterinary Microbiology. 2005; 105:29-36.
33 Murphy M, Buckley JF, Whyte P, Mahony MO, Anderson W, Wall PG, Fanning S. Surveillance of Dairy Production Holdings Supplying Raw Milk to the Farmhouse Cheese Sector for Escherichia coli O157, O26 and O111. Zoonosis and Public Health, 2014; 54:358-365.
34 Hussein HS, Sakuma T. (2005). Invited Review: Prevalence of Shiga Toxin-Producing Escherichia coli in Dairy Cattle and Their Products. Journal of Dairy Science. 2005; 88:450-465.
35 Pieri FA, Colombo M, Merhi CM, Juliati VA, Ferreira MS, Nero MA, Nero LA. Risky consumption habits and safety of fluid milk available in retail sales outlets in Viçosa, Minas Gerais State, Brazil. Foodborne Pathogens and Diseases. 2014; 6:490-496.
36 Ferreira MRA, Stella AE, Freitas-Filho EG, Silva TS, Nascimento KA, Pinto JFN, Dias M, Moreira CN. Distribution of the stx1 and stx2 genes in Escherichia coli isolated from milk cattle according to season, age and production scale in southwestern region of Goiás, Brazil. Arquivo Brasileiro de Medicina Veterinária e Zootecnia. 2018; 70(6):1807-1813.
37 Fremaux B, Raynaud S, Beutin L, Rozand CV. Dissemination and persistence of shiga-toxin producing Escherichia coli (STEC) on French dairy farms. Veterinary Microbiology. 2006; 117:180-191.
38 Murinda SE, Ibekwe AM, Rodriguez NG, Quiroz KL, Mujica AP, Osmon K. Shiga toxin-producing Escherichia coli in mastitis: an international perspective. Foodborne Pathogens & Diseases. 2019; 16(4). Doi: 10.1089/fpd.2018.2491
39 Ranjbar R, Dehkordi FS, Shahreza MHS, Rahimi E. Prevalence, identification of virulence factors, O-serogroups and antibiotic resistance properties of Shiga-toxin producing Escherichia coli strains isolates from raw milk and traditional dairy products. Antimicrobial resistance & Infection Control. 2018; 7(53).
40 Honish L, Predy G, Hislop N, Chui L, Kowalewska-Grochowska K, Trottier L, Kreplin C, Zazulak I. An Outbreak of E. coli O157:H7 Hemorrhagic Colitis Associated with Unpasteurized Gouda Cheese. Canadian Journal of Public Health. 2005; 96:182-184.
41 Denny J, Bhat M, Eckmann K. Outbreak of Escherichia coli O157:H7 Associated with Raw Milk Consumption in the Pacific Northwest. Foodborne Pathogens. 2008; 6:321-328.
42 Paneto BR, Schocken-Iturrino, RP, Macedo C, Santo E, Marin JM. Ocurrence of toxigenic Escherichia coli in raw milk cheese in Brazil. Arquivo Brasileiro de Medicina Veterinaria e Zootecnia. 2007; 59:508-512.
43 Vidal-Martins AMC, Bürger KP, Gonçalves ACS, Grisólio APR, Aguilar CEG, Rossi GAM. Evaluation of consumption of milk and its informal derivatives and the knowledge of the population about the public health problems in a municipal district in São Paulo State, Brazil. Boletim de Indústria Animal. 2013; 70:221-227.
44 Jaakkonen A, Castro H, Hallanvuo S, Ranta J, Rossi M, Isidro j, Lindström M, Hakkinen M. Longitudinal study of Shiga toxin-producing Escherichia coli and Campylobacter jejuni on finnish dairy farms and in raw milk. Applied and Environmental Microbiology. 2019; 85(7):e02910-18.