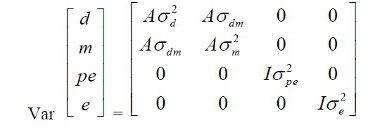
EFFECT OF SEVERAL STRUCTURES OF CONTEMPORARY
GROUPS ON ESTIMATES OF (CO)VARIANCE AND GENETIC PARAMETERS FOR WEANING WEIGHT
IN NELLORE CATTLE
Lillian Pascoa1, Arcadio de los Reyes2, Mauricio A. Elzo3, Jorge Luiz Ferreira4, Luiz A.F. Bezerra5, Raysildo Barbosa Lobo5
ABSTRACT
We used actual and adjusted weights
to 120 d and 210 d of age of 72,731 male and female Nellore calves born in 40
PMGRN - Nellore Brazil herds from
1985 to 2005 aiming to compare the effect of different definitions of
contemporary groups on estimates of
(co)variance and genetic parameters. Four models, each one with a different
structure of contemporary group (CG),
were compared using the Akaike Information Criterion (AIC), the Bayesian
Information Criterion (BIC), and the Consistent Akaike Information Criterion
(CAIC). (Co)variance estimates were obtained using a derivative-free restricted
maximum likelihood procedure. Estimates of (co)variances and genetic parameters
were similar for the four models considered. However, the BIC and CAIC
indicated that the most appropriate model for this Nellore population was the
one that considered CG to be random,
and sex of calf to be fixed and separate from CG, in which CG was
defined as the group of calves born in the same herd, year, season of birth
(trimester), and undergone the same management.
--------------------
EFEITO DE DIFERENTES MODELOS SOBRE AS
ESTIMATIVAS DE (CO)VARIÂNCIAS E PARÂMETROS GENÉTICOS PARA PESOS ATÉ A DESMAMA
EM GADO NELORE
RESUMO
---------------------
PALAVRAS-CHAVE: critérios de informação; gado de corte; grupos
contemporâneos.
INTRODUCTION
The
structure of contemporary groups (CG)
is of primary importance for genetic evaluation of animals under selection;
they are crucial to avoid potential biases in genetic evaluations due to
differential treatment of animals in a population (VAN VLECK, 1987).
Contemporary
groups have usually been considered as fixed effects in beef cattle genetic
evaluations. This has been based on HENDERSON (1973) statement that in sire models,
genetic predictions of sires would be associated to contemporary group effects,
and to eliminate this bias, CG needed to be defined as fixed effects.
Currently, the model of choice is an animal model where individuals are assumed
to represent a random sample of the genetic material in a population; however, CG continues to be considered as fixed
effects.
Some
authors have found that random CG
effects yield a better adjustment than models with fixed CG in various animal species. In small herds, BABOT et al. (2003)
managed to estimate genetic values for litter size in herds with insufficient
number of animals per CG using
simulated data, whereas VASCONCELOS et al. (2005) estimated genetic values for
milk production in dairy cattle in Portugal using contrast models. Treating CG as random effects was also found to
be advantageous by GONZÁLEZ-RECIO &
ALENDA (2005) when analyzing binary reproductive traits in Spanish dairy
cattle, by WOLF et al. (2005) for growth and litter size in swine
utilizing a multi-trait animal model, and by LEGARRA et al. (2005) for milk
production in ewes using a Bayesian approach.
To
obtain the best possible estimates of (co)variance and genetic parameters it is
important to define mathematical models that fit the available data as
accurately as possible. This will in turn yield the most accurate genetic
predictions given the available information. Thus, the objective of this study
was to compare models with different definitions of contemporary groups on
estimates of (co)variances and genetic parameters for actual and adjusted
weights at 120 and 210 d of age in Nellore cattle in Brazil.
MATERIAL AND METHODS
Actual and adjusted weights at 120 d (AW120,
RW120) and 210 d (AW210, RW210) from 72,731 male and female Nellore calves born
between 1985 and 2005 in 40 herds from PMGRN-Nellore Brazil were used in this
study. Actual weights were the closest ones to 120 d and 210 d within the
intervals of 120 ± 90 d and 210 ± 90 d, respectively. Calf ages were expressed
as deviations (CAD) from 120 d and 210 d. Adjusted weights were computed by
interpolation between a prior and a posterior weight to the standardized age
(120 d or 210 d), allowing a maximum interval of 195 d between these two
weights (± 90 d plus an additional 15 d due to possible management changes).
Birth weight was used as the prior weight for AW120 when there was no other
weight (actual birth weight or breed mean: 33 kg for males, and 31 kg for
females) to compute the interpolation. Computations were similar to PMGRN
(LÔBO, 1996):
The
effect of age of cow in years was grouped into six classes (DAC): 1 = 2 yr; 2 =
3 yr; 3 = 4 yr; 4 = 5 yr; 5 = 6 to 9 yr; and 6 = 10 yr and older cows.
Four
structures of contemporary groups were defined by concatenation of individual
effects, starting from a base subclass (CGB), as follows:
CGB:
herd – year of birth – management group at each age.
CG1: CGB – semester of
birth.
CG2: CGB – trimester of
birth.
CG3: CG1 – sex of calf.
CG4: CG2 - sex of calf.
Based on these four CGs (CG1 to CG4),
four analytical models were defined:
M1: Weight = α + CG1 + SC + DAC + ε
M2: Weight = α + CG2 + SC + DAC + ε
M3: Weight = α + CG3 + DAC + ε
M4: Weight = α + CG4 + DAC + ε
where,
Weight = actual or adjusted weight at 120 d or 210 d of age; α = constant; CG = contemporary
group; SC = sex of calf; DAC = class of cow age at calving, and ε = random residual effect. In
addition, models for actual weights included age of calf at weighing (CAD),
modeled as a cubic polynomial regression, and expressed as a deviation from 120
d or 210 d.
A minimum of five observations per contemporary
groups were required. Calves in CG with less than five observations were kept
in the database but their weights were set to zero, thus their genetic
evaluations were computed using solely information from their relatives. This
allowed us to have the same inverse of the relationship matrix (A-1)
with 119,586 animals in all analyses.
Models
for the estimation of (co)variances and genetic parameters for AW120, RW120,
AW210 e RW210 using single-trait analysis, considering CG fixed (1) or random
(2), were as follows:
y = Xb + Z1d + Z2m
+ Z3pe + e
(1)
y = Xb + Z1d + Z2m
+ Z3pe + Z4c + e (2)
where,
y = vector of observations; b = vector of fixed effects, including CG (Equation
1), and the effects of SC, DAC, and a cubic polynomial regression on CAD for
the analysis of actual weights (Equations 1 and 2); d, m, pe, c and e = vectors
of additive direct genetic effects, additive maternal genetic effects, maternal
permanent environmental effects, contemporary group, and residual, respectively;
and X, Z1, Z2, Z3 e Z4, are known
incidence matrices relating observations in vector y to vectors b, d, m, pe,
and c, respectively. The assumptions of
these models were:
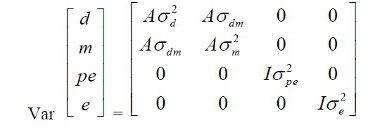
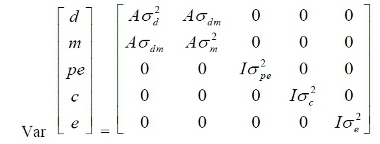
for
model 2, where, A = matrix of additive relationships among individuals; I = identity
matrices of appropriate order, σ²d, σ²m, σ²pe, σ²c and σ²e = additive direct genetic variance, additive
maternal genetic variance, permanent environmental variance, contemporary group
variance, and residual variance, respectively; and σdm= covariance
between direct and maternal genetic effects.
Models
were compared using the logarithm of the likelihood function (logL), using the
Akaike Information Criterion (AIC; AKAIKE, 1972), the Bayesian Information
Criterion (BIC; SCHWARZ, 1978), and Consistent Akaike Information Criterion
(CAIC, BOZDOGAN, 1987). The CAIC gives higher penalties to hyperparametrized
models compared to AIC. Thus, BIC and CAIC favor parsimonious models. These
criteria are defined as follows:
AIC = -2logL + 2k
BIC = -2logL + klog(n)
CAIC = -2logL + k(log(n)+1)
where,
k = number of estimated parameters; n = number of observations; logL =
logarithm of the likelihood function.
Models
with lower values of these information criteria are considered to better fit
the data.
Estimates
of (co)variances and genetic parameters were obtained using a derivative-free
restricted maximum likelihood procedure (DFREML; SMITH & GRASER, 1986). Computations
were carried out using the MTDFREML (Multiple Trait Derivative Free Restricted
Maximum Likelihood; BOLDMAN et al., 1995) software package using a single-trait
animal model. Because comparisons among animals were done within contemporary
groups, the variance due to CG, in
those models that considered CG to be random, was not included in the
phenotypic variance.
RESULTS AND DISCUSSION
Model Fitting
Values for the information criteria AIC, BIC and
CAIC obtained using models 1 through 4 with CG either fixed or random are
presented in Table 1 for 120 d and in Table 2 for 210 d of age. By any of these
criteria, better fitting models have smaller values.
In all cases, the criterion AIC had the smallest
values for models with CG fixed. On
the other hand, BIC and CAIC, by imposing higher penalties than AIC for models
with higher number of estimated parameters, favored models with random CG. These results were in agreement with
results from the literature. UGARTE et al. (1991), working with simulated data,
and VISSHER & GODDARD (1992), working with dairy data from small herds,
estimated lower prediction error variances (PEV) and mean squared errors (MSE)
for models with random CG. Contrarily, VALVERDE et al. (2008), using Braunvieh
cattle weaning weight data, found somewhat higher accuracies of genetic
predictions for direct genetic effects when CG were considered fixed, and no
difference between models with CG fixed or random for maternal genetic effects.
For weights at 120 d (AW120 and RW120), the criteria
BIC and CAIC indicated that models with trimester seasonal effect had the best
fit when CG was random (Table 1). A similar result was obtained for weights at
210 d (AW210 and RW210; Table 2). However, when CG was fixed, the best fitting
models were those with semester seasonal effects for 120 d (AW120 and RW120),
whereas for weights at 210 d, model M2 (trimester) was better than M1
(semester), but model M3 (semester) was better than M4
(trimester) for AW210 and RW210. On the other hand, REYES et al. (1998) found
out that it was better to consider trimester over semester when they compared
the efficiency of two fixed effects models containing season (trimester or
semester) as part of the structure of contemporary groups for weaning weight in
Nellore cattle.
According to the BIC and CAIC criteria, models that
had sex of calf effect separated from CG, provided a better fit to the data.
These models allowed the construction of contemporary groups with larger number
of individuals and permitted better genetic connections among CG and higher
accuracies of prediction of genetic evaluations. These results are in agreement
with those found by REYES
et al. (2006) for growth between birth and weaning in a multibreed Nellore x
Hereford cattle population.
Among models with random CG, model M2 was
the most parsimonious and provided the best fit. Thus, for 120 d and 210 d of
age and actual or adjusted data, results here suggest that models that had
random CG with trimester seasonal effect, and sex of calf effect separated from
CG effect were the most appropriate for growth from birth to weaning in Nellore
cattle.
Tables 3 and 4 present the estimates of
(co)variances and genetic parameters for the four traits in this study (AW120,
RW120, AW210 e RW210). Estimates for each trait differed little among models.
Estimates of σ²d, σ²m, h²d and h²m were larger in models with semester of birth
season effect in CG (M1 and M3) than those obtained in
models with trimester of birth season effect in CG (M2 e M4).
On the other hand, estimates of σ²pe and σ²e were similar in all
models. These results may have been due to greater variation among weights when
the period of time (season effect) considered in CG was longer.
Estimates of σ²d, σ²m, h²d and h²m for weight at 120 d and 210 d of age were similar in models that included or not the
effect of sex of calf within CG (M2 vs M4 and M1
vs M3). Estimates of σ²e ranged from 118.61 to 163.01 kg2 for
actual and adjusted weights at 120 d (W120), and from 223.99 to 292.43 kg2
for actual and adjusted weights at 210 d (W210), with smaller values in
CG of larger size, i.e., those in models that considered semester seasonal
effects and sex of calf separately from CG.
Models that considered CG random yielded higher
estimates of σ²d, σ²m, h²d and h²m , and lower estimates
of σ²e than models that considered CG fixed. Larger
estimates of σ²d and smaller values of σ²e in models with random CG were also obtained
for weaning weights in Braunvieh cattle in México (VALVERDE et al., 2008).
Literature values reported higher values of heritability estimates for models
with fixed CG than for models with random CG (PHOCAS & LALOE, 2003;
CHANVIJIT et al., 2005; VALVERDE et al., 2008). However, these lower heritability
estimates for models with random CG were computed with phenotypic variances
that included the variance due to contemporary group (σ²c), which is not
appropriate because comparisons among animals evaluated genetically occur
within contemporary groups.
Estimates for σ²d ranged from 56.20 to 68.17 kg2 and
estimates for σ²m from 24.86 to 29.67 kg2 for weight
at 120 d of age (W120). For weights at 210 d of age (W210), σ²d estimates ranged from 116.61 to 155.44 kg2
and estimates of σ²m ranged from 41.91 to 52.05 kg2.
Estimates of σ²dm were negative, ranging from -38.61 to -20.84 kg2 for W120 and
from -38.94 to -21.41 kg2
for W210. These estimates indicated antagonism between additive direct and
maternal genetic effects, in agreement with previous beef cattle research
(FERREIRA et al., 1999; LEE & POLLAK, 2002; ROSALES et al., 2004). This
implies that if producers perform selection for growth in calves without
considering maternal additive genetic effects, this may produce a decrease in
milk production of future mothers and a reduction in weaning weights of their
progenies (VALVERDE et al., 2008).
Estimates of σ²pe ranged from 30.38 to 33.97 kg2 for
W120 and from 66.59 to 74.27 kg2 for W210. These estimates were
higher than those reported by GARNERO et al. (2001) for weights at 120 d of age
(19.1 kg2) and at 220 d of age (48.01 kg2).
Direct heritability estimates ranged from 0.22 to
0.28 for W120 and from 0.25 to 0.32 for W210. Estimates of maternal
heritabilities ranged from 0.10 to 0.12 for W120 and from 0.09 to 0.11 for
W210. MARCONDES et al. (2002) and SIQUEIRA et al. (2003) found similar
estimates of direct heritability for W120 (0.24 and 0.29, respectively), and of
maternal heritability (0.08) in Nellore cattle. GARNERO et al. (2001) also
estimated values of heritability for direct genetic effects (0.19) and for
maternal genetic effects (0.06) similar to those obtained here.
Differences among estimates of (co)variances and
genetic parameters were small across models in this study, perhaps due to the
utilization of the same matrix of additive relationships. However, one could
expect that estimates from models that yield better fit would be more accurate
and reliable.
CONCLUSIONS
REFERENCES
AKAIKE, H. Information theory and an extension
of maximum likelihood principle. In: INTERNATIONAL SYMPOSIUM ON INFORMATION
THEORY, 2., Proceedings… p.267-281,
1972. Disponível em http://www.sortiend.org/lme/Statistical%20Papers/Akaike_1973%20with%20commentary.pdf, acesso em 03 de março de 2013.
BABOT, D.; NOGUERA, J.L.; ALFONSO, L.; ESTANY,
J. Fixed or random contemporary groups in genetic evaluation for litter size in
pigs using a single trait repeatability animal model. Journal of Animal Breeding and Genetics, v. 120, n. 1, p.12-22,
2003.
BOLDMAN, K.; KRIESE, L.; VAN VLECK, L.D. A manual for use of MTDFREML : A set of
programs to obtain estimates of variances and covariances. USDA. ARS., 1993.
BOZDOGAN, H. Model selection and Akaike’s
information criterion (AIC): the general theory and its analytical extensions. Psychometrika, v. 52, p.345-370, 1987.
CHANVIJIT, K.;
DUANGJINDA, M.; PATTARAJINDA, V.; REODECHA, C. Model comparison for genetic
evaluation of Milk yield in crossbred Holsteins in the tropics. Journal of
Applied Genetics, v. 46, n. 4, p.387-393, 2005.
FERREIRA, G.B.;
MACNEIL, M.D.; VAN VLECK, L.D. Variance components and breeding values for
growth traits from different statistical models. Journal of Animal Science,
v. 77, p. 2641-2650, 1999.
GARNERO, A.V.; LOBO, R.B.; BEZERRA, L.A.F.;
OLIVEIRA, H.N. Comparação entre alguns critérios de seleção para crescimento na
raça Nelore. Revista Brasileira de Zootecnia, v.30, n. 3, p.714-718, 2001.
GONZÁLEZ-RECIO, O.;
ALENDA, R. Genetic parameters for female fertility traits and a fertility index
in Spanish dairy cattle. Journal of Dairy Science, v. 88, p. 3282-3289,
2005.
HENDERSON, C.R. Sire evaluation and genetics
trends. In: ANIMAL BREEDING AND GENETIC SYMPOSIUM IN HONOR OF DR. JAY L. LUSH, Illinois, USA. Proceedings… Illinois, v. 1, p.10-41,
1973. Disponível em: <http://jas.fass.org/cgi/reprint/1973/Symposium/10> Acesso em: 15 mar.
2011.
LEE, C.; POLLAK, E.J.
Genetic antagonism between body weight and milk production in beef cattle. Journal
of Animal Science, v. 80, p.316-32, 2002.
LEGARRA, A.; LÓPEZ-ROMERO, P.; UGARTE, E.
Bayesian model selection of contemporary groups for BLUP genetic evaluation in
Latxa dairy sheep. Livestock Production Science, v. 93, p.205-212, 2005.
LÔBO,
R.B. Programa de Melhoramento Genético
da Raça Nelore, 2 Ed. Ribeirão Preto, SP: ANCP, 1996.
MARCONDES, C.R.; GAVIO, D.; BITTENCOURT, T.C.C.;
LÔBO, R.B.; BEZERRA, L.A.F.; TONHATI, H. Estudo de modelo alternativo para
estimação de componentes de (co)variância e predição de valores genéticos de
características de crescimento de bovinos da raça Nelore. Arquivo Brasileiro
de Medicina Veterinária e Zootecnia, v. 54, n. 1, p. 93-99, 2002.
PHOCAS, F.; LALOË, D. Evaluation models and genetic parameters for calving difficulty in beef
cattle. Journal
of Animal Science, v. 81, p.933-938, 2003.
REYES,
A.; LOBO, R.B.; BEZERRA, L.A.F.; MARTINS, E.N. Critérios de época na conformação
de grupos contemporâneos para características de crescimento em Nelore. In: REUNIÃO ANUAL DE SOCIEDADE BRASILEIRA DE
ZOOTECNIA, 35., Botucatu. Anais... Botucatu : SBZ, v. 3, n. 1, p. 437-439, 1998. Disponível em http://www.sbz.org.br/reuniaoanual/anais/arq_reuniao_anual/sbz1998.rar. Acesso em 03 março 2013.
REYES, A.; ELZO, M.A.; ROSO, V.M.; FRIES, L.A.;
CARVALHEIRO, R. Efeitos não genéticos
na composição dos grupos contemporâneos para o ganho de peso do nascimento a
desmama de uma população multirracial Nellore x Hereford. In: REUNIÃO ANUAL DA SOCIEDADE BRASILEIRA DE
ZOOTECNIA, 43., João Pessoa. Anais... João Pessoa, 2006. Disponível em:
<http://www.animal.ufl.edu/elzo/Publications/Proceedings/2006/Sociedad%20Brazileira%20de%20Zootecnia/Reyes%20et%20al._
SBZ43_July-24-27-2006_Trabalho_HOMEPAGE.pdf. Acesso em 20 out. 2009.
ROSALES, A.J.; ELZO, M.A.; MONTANO, B.M. Parámetros y tendencias
genéticas para características de crecimiento predestete em la población
mexicana de Simmental. Revista Mexicana de Ciencias Pecuarias, v. 42, p.
171-180, 2004.
SCHWARZ,
G. Estimating the dimension of a model. The Annals of Statistics, v.
6, n. 2, p.461-464, 1978. Disponível em: < http://www.jstor.org/stable/2958889> Acesso em: 15 mar.
2011.
SIQUEIRA, R.L.P.G.; OLIVEIRA, J.A.; LOBO, R.B.;
BEZERRA, L.A.F.; TONHATI, H. Análise da variabilidade genética aditiva de
características de crescimento na raça Nelore. Revista
Brasileira de Zootecnia,
v. 32, n. 4, p. 880-886, 2003.
SMITH, S.P.; GRASER, H.U. Estimating Variance Components in a Class of
Mixed Models by Restricted Maximum Likelihood. Journal of Dairy Science, v. 69, p. 1156-1165, 1986.
UGARTE, E.; ALENDA,
R.; CARABAÑO, M.J. Fixed or random contemporary groups in genetic evaluations. Journal of Dairy Science, v. 75, p. 269-278, 1991.
VALVERDE, R.R.;
DOMÍNGUEZ, R.N.; FLORES, A.R.; MUÑÍS, J.G.G.; VALENCIA, F.M. Comparación de
definiciones de grupos contemporáneos en la evaluación genética de bovinos
Suizo Europeo. Revista Mexicana de Ciencias Pecuarias, v. 46, n. 4, p. 359-370, 2008.
VAN VLECK, L.D. Contemporary Groups for Genetic Evaluations. Journal of Dairy Science,
v. 70, p. 2456-2464,
1987.
VASCONCELOS,
J.; MARTINS, A.; FERREIRA, A.; CARVALHEIRA, J. Consequências da eliminação de
rebanhos pequenos da avaliação genética de bovinos leiteiros em Portugal. Revista Portuguesa de
Zootecnia, v. 12, p. 105-117, 2005.
VISSHER, P.M.;
GODDARD, M.E. Fixed and random contemporary groups. Journal of Dairy Science,
v. 76, p. 1444-1454, 1992.
WOLF, J.; ZAKOVA, E.; GROENEVELD, E. Genetic parameters for a joint genetic evaluation of production and reproduction traits in pigs. Czech Journal of Animal Science, v. 50, n. 3, p. 96-103, 2005.